Snippets from my research (non-chronological)
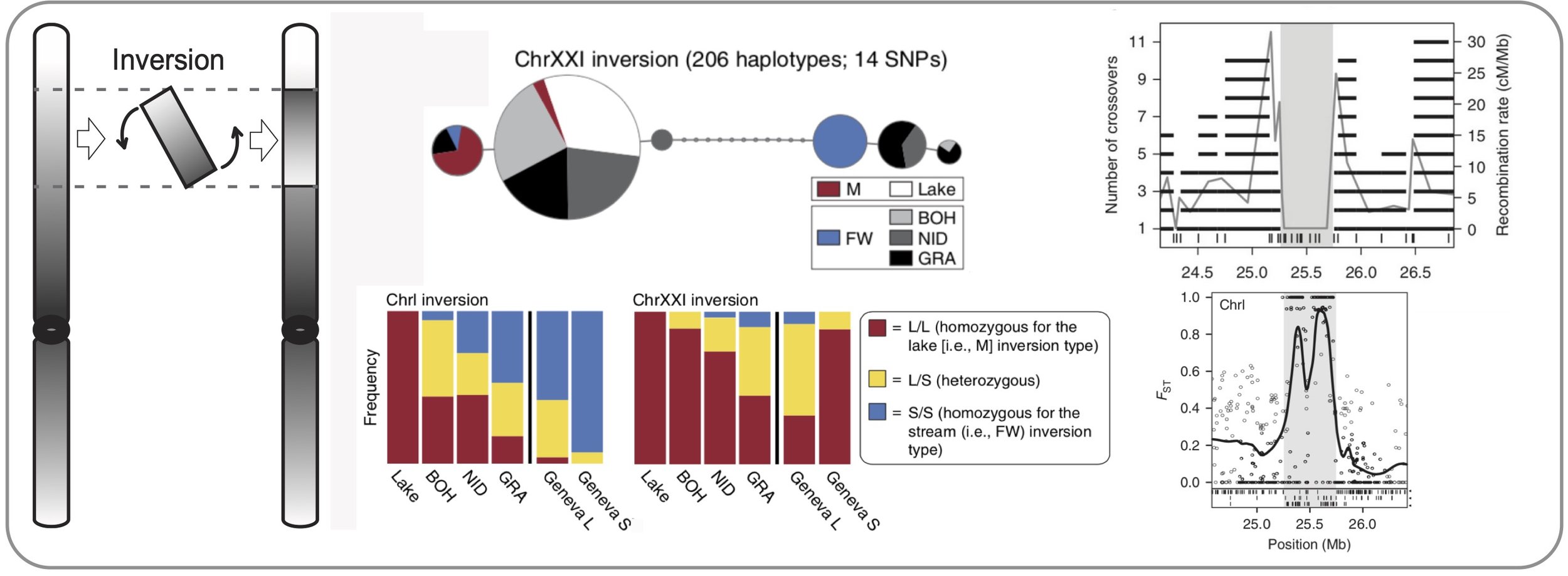
Inversions & adaptation. We show that old chromosomal inversions differentiate parapatric lake and stream stickleback in a Central European watershed. Because lake-stream differentiation at these inversions occurs repeatedly, the inversions are likely important for adaptive divergence. We show experimentally that recombination between the two inversion variants is absent. However, genomic differentiation between individuals carrying different variants suggests that rare recombination events – such as double crossovers or gene conversion – have occurred in the center of the inversions. (Roesti et al. 2015)
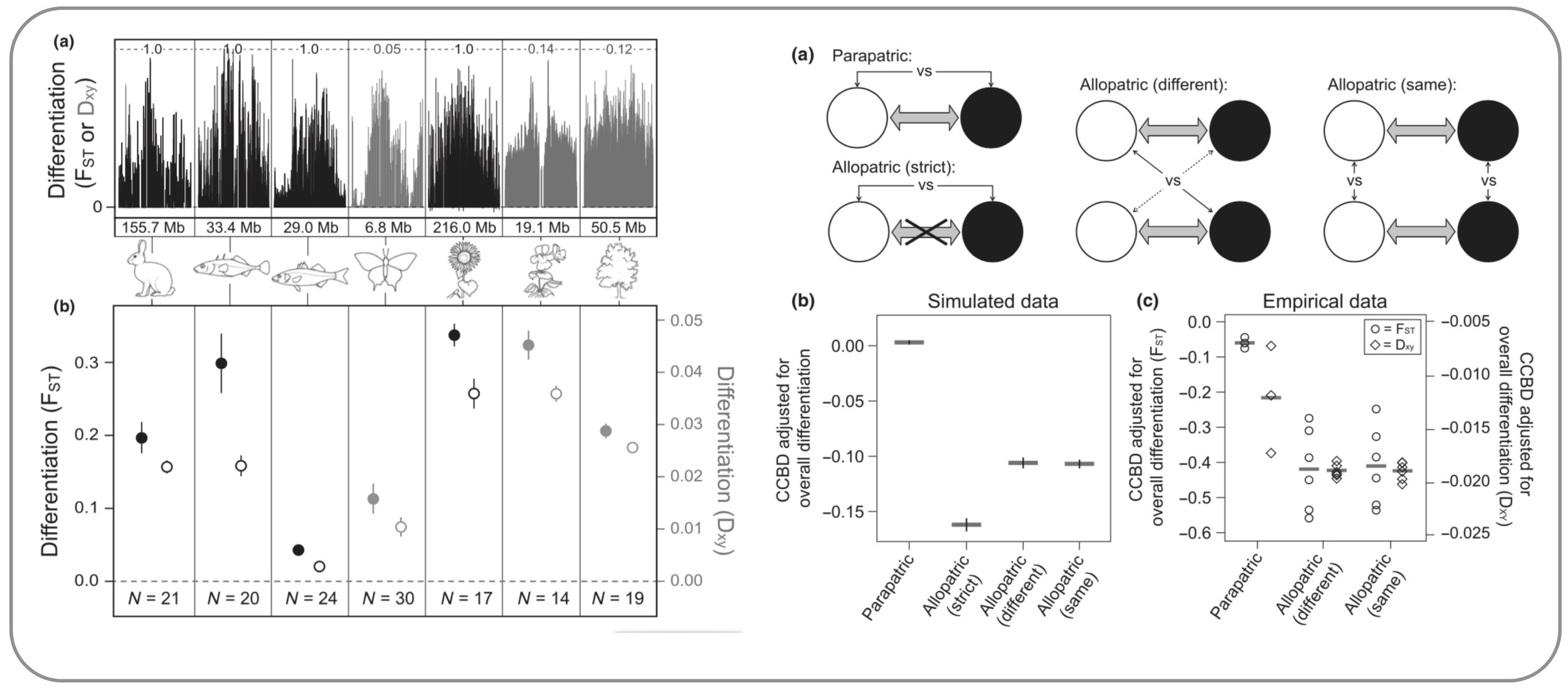
Recombination strongly influences genome-wide population divergence. Chromosome-scale patterns of genomic differentiation are similar across many organisms, with higher recombination at chromosome tips and lower recombination in chromosome centers. This variation in recombination, together with polygenic selection and gene flow, strongly shapes genomic patterns of divergence. In addition to describing this empirical pattern, we use simulations to quantify different genetic mechanisms that lead to elevated population divergence in regions of low recombination, including genetic hitchhiking and barriers to gene flow. We then test the predictions from these models by comparing allopatric and parapatric populations of stickleback residing in different or similar environments. (Berner & Roesti 2017)
Species divergence driven by resource competition and shared predation. By comparing co-occurring (sympatric) and allopatric populations of both threespine stickleback and prickly sculpin fish, we find that trait shifts occur in opposite directions between the species when they are sympatric. These shifts are evident in typical trophic traits and diet, as well as in traits that provide protection against large predators. Our study suggests that ecological character displacement, driven by resource competition, has increased the vulnerability of stickleback but decreased the vulnerability of sculpin to a shared predator (trout). This highlights the importance of indirect interactions between prey species – mediated by shared predators – in driving species divergence. (Roesti et al. 2022)
Genomic architecture of adaptation with gene flow. In this perspective, I summarize why and how adaptive loci are expected to cluster within the genome when local adaptation occurs in the presence of maladaptive gene flow. I focus in particular on the role of low-recombination regions in promoting such clustering. Despite growing interest in this topic, we still have a limited understanding of where adaptive loci are located in the genome. Although low-recombination regions appear to be promising hotspots for clustering, observed patterns may often have alternative explanations, including recombination-related biases in our genomic methods. I illustrate this bias using hundreds of previously published QTLs in threespine stickleback and suggest future directions to address these challenges through comparative genomics. (Roesti 2018)
Chromosomal inversions as a constraint to new adaptation. In this perspective, we reason and use simulations to illustrate that chromosomal inversions can limit adaptation to new habitats because inversions limit the reshuffling of existing genetic variation into newly favorable combinations. (Roesti et al. 2022)
The largest test of the Biotic Interactions Hypothesis. It is widely assumed that species interactions, such as predation, intensify from the poles toward the equator and may help explain the higher species diversity found in the tropics. We tested this so-called “Biotic Interactions hypothesis” in the open ocean using 55 years of catch-per-effort data from pelagic longline fishing. Surprisingly, we found that predation by large open-water fish is strongest at temperate latitudes, and predation is negatively correlated with species richness. These findings run counter to the predictions of the biotic interactions hypothesis. (Roesti et al. 2020)
A function trade-off between foraging and brood care – and not sexual selection – explains sexual dimorphism in cichlids. Cichlid fish species in East Africa’s Lake Tanganyika differ in their modes of parental care: in some species, only one sex or both sexes perform mouthbrooding, while in others, parental care involves no mouthbrooding at all. We predicted that, due to the likely dual function of gill rakers in both foraging and mouthbrooding, only species with uniparental mouthbrooding would show sexual dimorphism in gill raker morphology. Indeed, this is what we found. Our study offers a largely overlooked explanation for sexual dimorphism in nature – one that cannot be accounted for by sexual selection or by initial niche divergence between the sexes. (Ronco*, Roesti*, Salzburger* 2019)
A simple biotic change leads to strong genome-wide adaptation in stickleback. In many postglacial lakes in Western Canada, threespine stickleback and prickly sculpin co-occur and interact through mainly food competition and via shared predators. In some lakes, however, only stickleback are present, with no sculpin. We find strong and parallel genomic signatures of selection in stickleback associated with the presence or absence of sculpin. Moreover, the extent of phenotypic and genomic adaptive divergence is positively correlated. This study highlights the importance of indirect species interactions in driving evolutionary (genomic) diversification. (Miller, Roesti, Schluter 2019)
Left: Parallel adaptation from shared genetic variation, such as from pre-existing (standing) or introgressed variation, produces a distinct genetic signature within a genome. We predict this signature using simulation models, and then confirm it using genome-wide and targeted sequencing of natural stickleback populations. (Roesti et al. 2014)
Right: Chromosome-wide variation in recombination rate shapes differentiation and diversity within the stickleback genome. Recombination rate is consistently elevated towards the tips of chromosomes, and reduced in the center of chromosomes – a pattern that appears to be unrelated to the position of the centromeres, but instead, is related to functional constraints during meiosis. We further find that recombination rate variation influences nucleotide composition within a genome. (Roesti et al. 2013)
Field work and experiments. Fieldwork is a central part of my research, and I have recently developed a growing interest in conducting manipulative field experiments to better test causal relationships in eco-evolutionary processes.
Genomic signatures of a morphological adaptation. Lake-stream divergence of stickleback in lateral plating and the associated molecular signatures (Roesti et al. 2015).
Phenotypic divergence vs. genomic divergence across several lake-stream stickleback populations. We studied stickleback from lakes and adjacent streams to characterize phenotypic traits and genome-wide signatures of habitat-specific selection. Our results reveal that gene flow constrains genome-wide differentiation between diversifying populations, and that the degree of phenotypic and genomic divergence is positively correlated. This study provides one of the first genome-wide demonstrations of how variation in recombination rate shapes genomic differentiation during diversification. (Roesti et al. 2012)
Left: Phenotypic plasticity or genetic differentiation? In this study, we found evidence for strong genome-wide differentiation between distinct lamprey ecotypes that were previously thought to be the product of phenotypic plasticity (Mateus et al. 2013).
Right: Phylogenomics of an adaptive radiation. Patagonotothen icefish species reveals incomplete species boundaries in this adaptive radiation (Ceballos et al. 2019).
What is limiting our ability to predict evolution? I here outline the two possible explanations: limited knowledge and the inherently unpredictable influence of stochasticity in evolution. (Roesti 2021)